Eco friendly farming – how bacterial biofertilisers can help increase agricultural sustainability
Written by Carl Pille, PhD Candidate
While modern agriculture requires agrochemicals (fertilisers, pesticides etc.) to maximise the yield of produce and ensure food availability, they can be damaging to the environment by releasing greenhouse gases or causing algal blooms in waterways. Biofertilisers comprised of plant beneficial microorganisms are a promising tool to increase agricultural sustainability. When we talk about biofertilisers we’re often referring to plant beneficial rhizobacteria (PBR). These bacteria reside on or around plant roots (hence rhizo) and confer various beneficial effects to its plant host, such as increasing nutrient availability, increasing stress resistance or stimulating a plants hormone system for increased growth and yield.
A large barrier to their implementation in farms is the inconsistent performance of PBR, mainly due to competition with microbes already present in the soil for the nutrient rich habitat surrounding a plant’s roots. Much like the local surfers in a recently gentrified beach town chasing tourists off their favourite waves, indigenous microbes don’t take too kindly to recently added PBR encroaching on their turf and quickly bully them out of existence (Figure 1).

Figure 1: Bacteria in biofertilisers are often outcompeted for resources by bacteria already residing in the soil. (Figure generated with Microsoft Copilot).
Traditionally, PBR have been selected for inclusion in biofertiliser formulations by growing them on various types of petri dishes to test for common plant beneficial traits. These include the ability to make insoluble phosphate available to plants or to fix gaseous nitrogen from the atmosphere to a liquid, plant available form (Figure 2). However, these methods give us a limited view of the potential of biofertiliser candidates and may overlook PBR with less common or unknown beneficial traits.
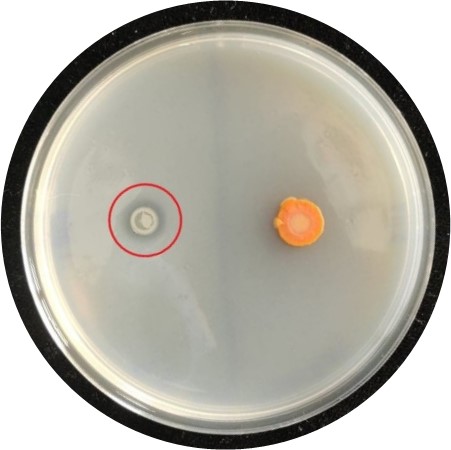
Figure 2: Two bacteria growing on a petri dish containing opaque, insoluble, plant unavailable phosphate, a critical plant nutrient. While not observed around the negative control bacteria (right, orange), a zone of clearing surrounding the bacterial colony on the left (red circle) indicates its ability to solubilise phosphate, making it available to plants. This is a traditional screen used to shortlist PBR.
In our study lead by myself, Carl Pille, at the University of Melbourne, we asked whether there was a way to select novel PBR with less common plant growth promoting traits which are less likely to be outcompeted by endemic microbes already in the soil. To do this we used an approach termed “bacterial metabarcoding”. This technique involves targeting a specific gene (or barcode) unique to each bacteria to gain a snapshot of an entire group of bacteria, or microbiome.
Using this approach, we were able to define a core set of bacteria which consistently colonised a model grass plant’s roots across distinct soil types and microbiomes. Interestingly, we were also able to isolate a subset of this core microbiome and found that several did not display any beneficial traits we screened for but still promoted plant growth (Figure 3). This highlights that the additional selection criteria afforded to us through the metabarcoding approach allows for detection of novel biofertiliser candidates with uncommon or new plant beneficial traits. These new bacteria may stand a better chance of persisting and surviving in the field, making them promising new biofertiliser candidates.

Figure 3: Some core root bacteria (Bacillus BD33 and Rhodococcus BD157) promoted plant growth compared to the control but did not display commonly screened for plant beneficial traits. This indicates that these bacteria have new or uncommon ways of promoting plant growth.
To read in details about this topic:
Click here for the journal article by Carl Pille et al.,
Learn more about Theme 3 research here.




